6.2 EMP_cluster_analysis
Cluster analysis falls under the domain of unsupervised learning in machine learning. It does not require predefined groups, instead, it investigates classification methods based on the inherent characteristics of the data and applies these methods to categorize the data reasonably. The result is grouping similar data into one set, which helps users uncover patterns and structures within the data, and identify outliers.
6.2.1 Cluster analysis of samples based on assay
🏷️Example:
MAE |>
EMP_assay_extract(experiment = 'geno_ec') |>
EMP_cluster_analysis()
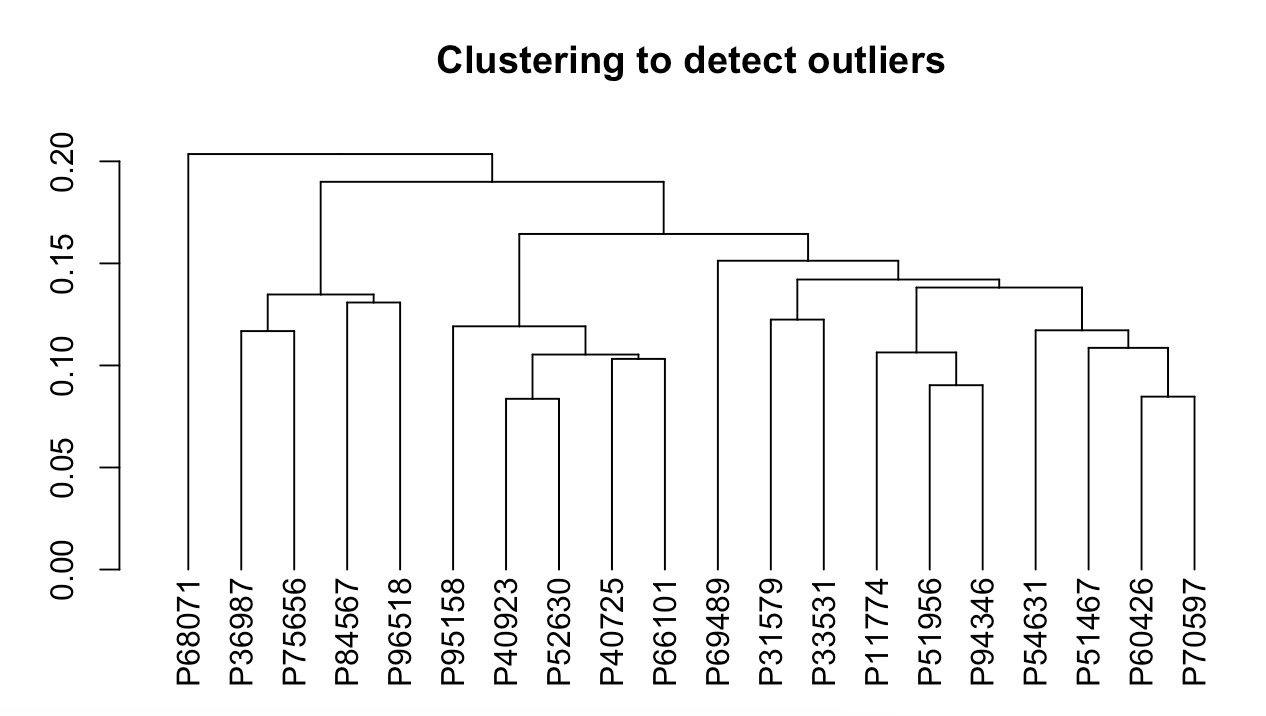
Clustering grouping tags can be specified through the parameter h.
MAE |>
EMP_assay_extract(experiment = 'geno_ec') |>
EMP_cluster_analysis(h=0.15)
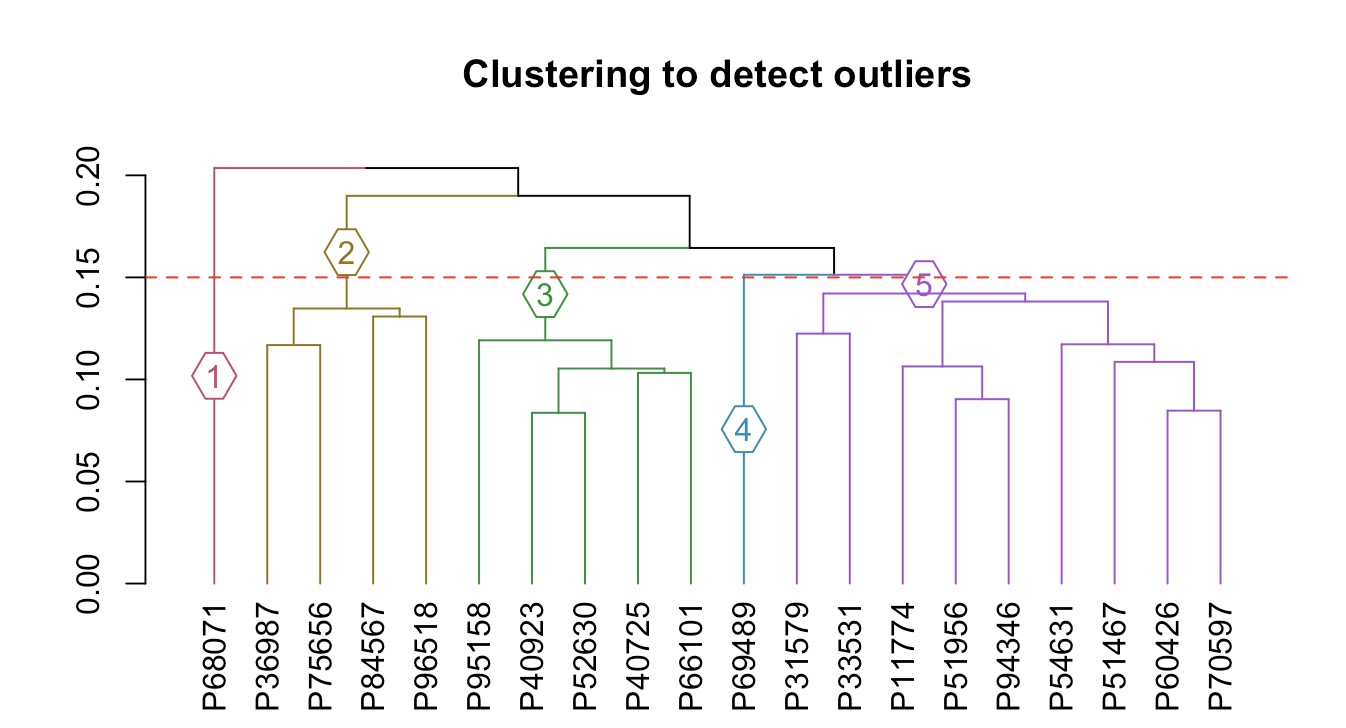
Use module EMP_filter to filter outliers in groups 1 and 4.
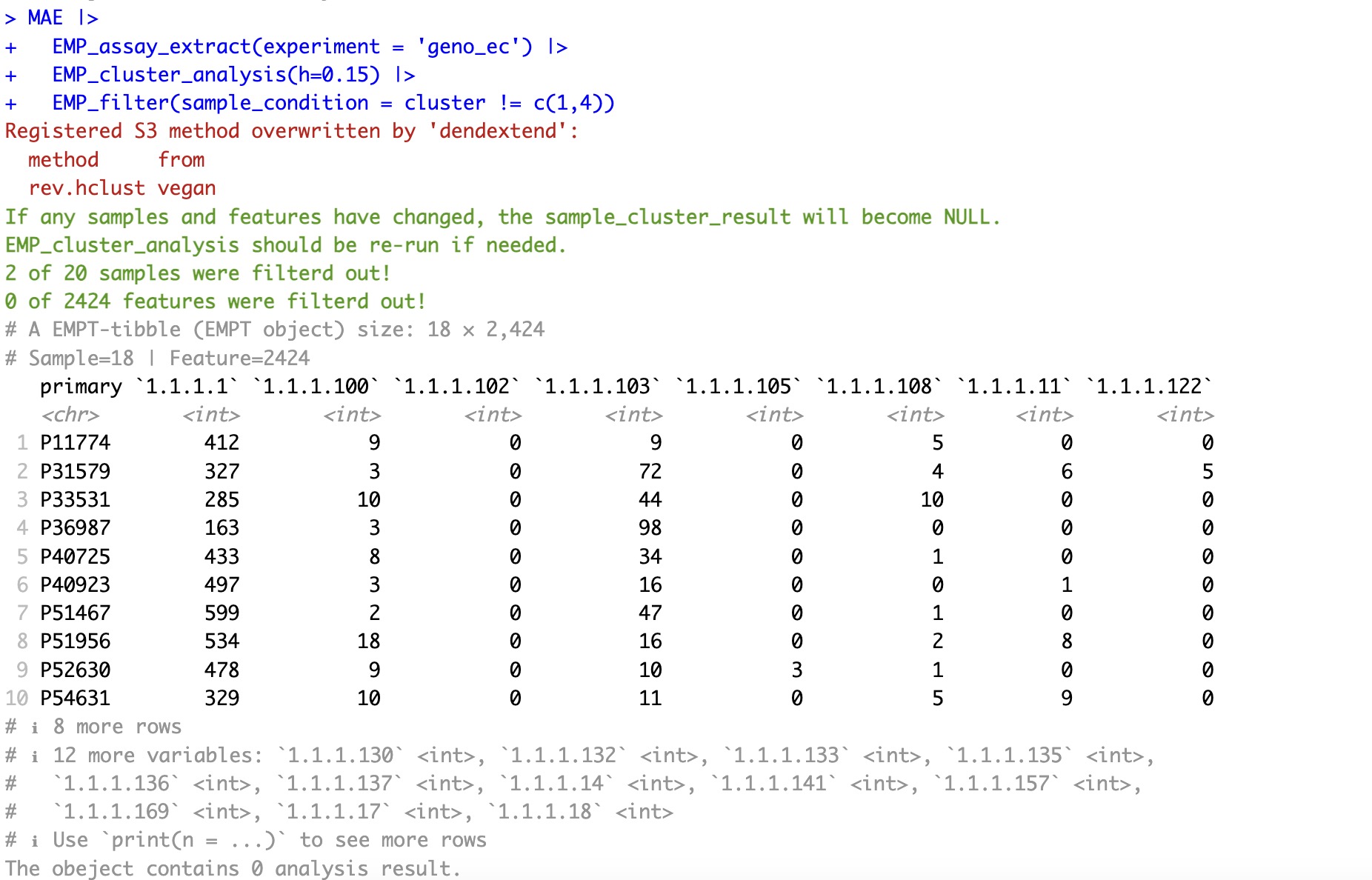
MAE |>
EMP_assay_extract(experiment = 'geno_ec') |>
EMP_cluster_analysis(h=0.15) |>
EMP_filter(sample_condition = cluster != c(1,4))
6.2.2 Cluster analysis of features based on assay
Clustering analysis can be performed on features to quickly discover hierarchical relationships among interesting features.
🏷️Example: Search for alcohol-related KO genes, and perform cluster analysis on the features.
This example identifies KO genes containing alcohol through searching for strings in the Name column of rowdata, rather than determining genes related to alcohol based on a knowledge graph.
 ### 6.2.3 Cluster analysis of samples based on coldata
In clinical data analysis, the sample-related data of the subjects typically includes basic information (such as dietary data, scale data, etc.). Module `EMP_colda_extract` can convert coldata into assay by specifying parameter `action='add'` , thus enabling downstream analysis.
🏷️**Example:**
### 6.2.3 Cluster analysis of samples based on coldata
In clinical data analysis, the sample-related data of the subjects typically includes basic information (such as dietary data, scale data, etc.). Module `EMP_colda_extract` can convert coldata into assay by specifying parameter `action='add'` , thus enabling downstream analysis.
🏷️**Example:**
①When there is a large amount of sample-related data in coldata, users can use the parameter
coldata_to_assay to select the relevant sample-related data for conversion. If not specified, all continuous variables in coldata will be converted to assay by default.②When the distance between some samples is missing due to missing values in certain features, the maximum default value of 1 will be used automatically. Users can modify the default value using the parameter
pseudodist, or use the module EMP_impute to impute missing values before clustering analysis.
MAE |>
EMP_coldata_extract(action = 'add',
coldata_to_assay = c('SAS','SDS','HAMA','HAMD','PHQ9','GAD7')) |>
EMP_cluster_analysis()
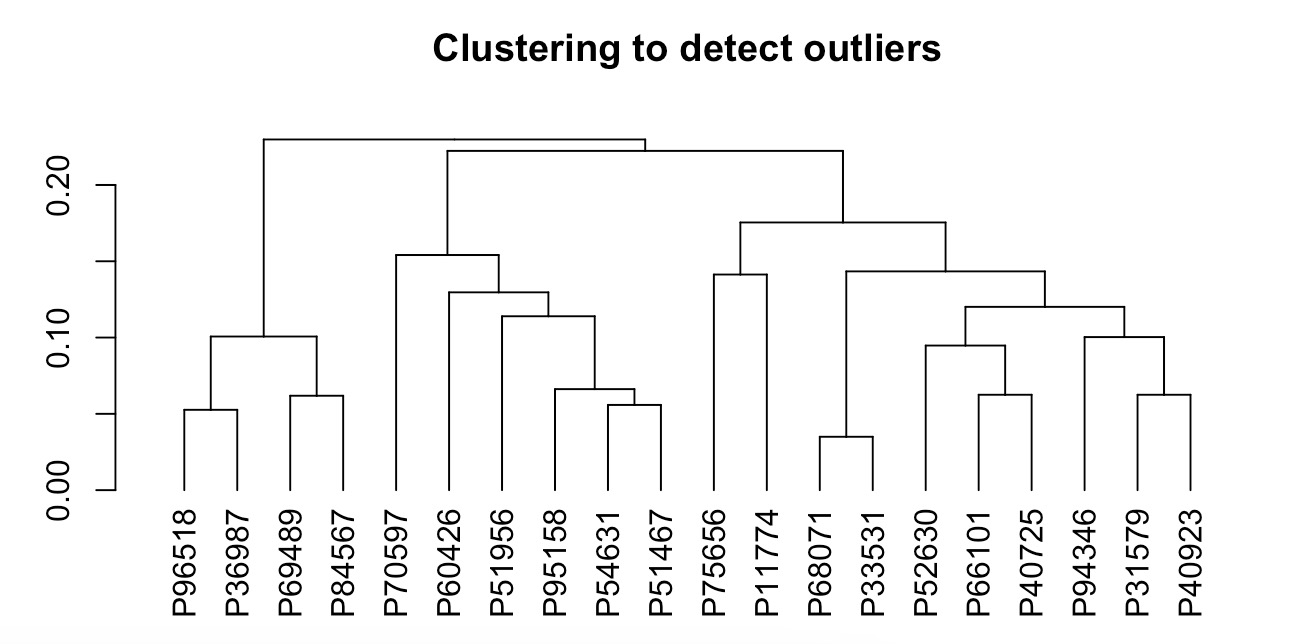
MAE |>
EMP_coldata_extract(action = 'add',
coldata_to_assay = c('SAS','SDS','HAMA','HAMD','PHQ9','GAD7')) |>
EMP_cluster_analysis(rowdata=TRUE)
